The most recent ctDNA paper verifies
its utility as a “broadly applicable, sensitive, and specific biomarker”
across “multiple different types of cancer” and useful in cancer
research and medicine: Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies
by Bettegowda et al Sci Transl Med 2014.
Some comments and analysis from the exciting and fast moving world of Genomics. This blog focuses on next-generation sequencing and microarray technologies, although it is likely to go off on tangents from time-to-time
Pages
▼
Monday, 24 February 2014
Saturday, 15 February 2014
The best talk at ABGT so far
This years AGBT has again been a full program from dawn to dusk (or later). This year my favourite session has been Fridays Genomic Technologies Development session. It was the most technologically focused session with five great talks and one that I thought was outstanding.
Friday, 14 February 2014
Everything you need to know about NGS library prep...almost
Jacques Retief (Sr. Manager Scientific Affairs at Illumina) has produced a phenomenal poster with graphics for over NGS 50 methods. If you've ever wanted to know how MAINE-seq, smMIP, ATAC-seq or just about anything else worked then Jaques helpfully provides a graphic of each method and the reference as well.
An image of the whole poster here would not do it sufficient justice so I'll stick to a few of the graphics and a link. Go get it, print it out and hang it on your bedroom wall!
Thursday, 13 February 2014
AGBT Wednesday 12th: No blogging allowed!
There were three great talks in the opening session of AGBT yesterday but before we kicked off Eric Green reminded everyone in the audience about the "opt-in" policy on blogging and tweeting (and any other kind of media). If someone opts out then we can't cover their talks.
AGBT speakers choosing to opt-out appears to fly in the face of what the major journals are aiming to do: keep the science community talking about science and not be overly restrictive. See what these journals have to say NEJM, Nature, Nature Methods, PLoS, Science, Cell.
Wednesday, 12 February 2014
Floridian roadkill on the way to AGBT
I had a long and tiring trip to Miami yesterday with two hours of immigration and security. I wok up at 4am thanks to jet-lag and decided to drive over to AGBT at Marco Island via I41 through the everglades and Big Cyprus national park. The weather was fantastic, especially given we've had months of rain and no sun in the UK. And the wildlife was fantastic.
Unfortunately so was the roadkill.
Unfortunately so was the roadkill.
Tuesday, 11 February 2014
Bloggers at AGBT, and blogs about bloggers...
AGBT is not all hard work; there's blogs to be written, Tweets to tweet and drinks to be drunk. Coverage from Bloggers and Tweeters is usually pretty good and makes up a little for not actually getting there, I know as I missed last year and caught up online.
Keep an eye on #AGBT and #notAGBT, as well GenomeWeb of course!
I just discovered The Science Web who are "putting the "omic" into comical". I'd recommend it if you want a light-hearted look at science. Apparently next year we'll all be gathering in North Korea! And they've a follow-up on Crap-Bio's SHT-seq system which we've been waiting for for far too long. Another recent post over at pokes fun at the science blogging community: Science blogging community makes contact with the outside world, we'll have to get our own back!
I just discovered The Science Web who are "putting the "omic" into comical". I'd recommend it if you want a light-hearted look at science. Apparently next year we'll all be gathering in North Korea! And they've a follow-up on Crap-Bio's SHT-seq system which we've been waiting for for far too long. Another recent post over at pokes fun at the science blogging community: Science blogging community makes contact with the outside world, we'll have to get our own back!
See you there, it's a shame it's going to rain for the next two days!
Sunday, 9 February 2014
Wet-lab, dry-lab or moist-lab
Wet-lab: your traditional research lab with lots going on at the bench (often collaborating with dry-labs).
Dry-lab: A computational or mathematical lab focusing on analysis of other peoples data (sometimes collaborating with dry-labs).
Moist-lab: the new style of lab with both wet- and dry-staff, working together with some post-docs now being truly multi-functional developing methods in both environments.
 |
| What do you call a moist-lab worker? |
Florian Markowetz is a junior group leader at the Cambridge Institute and gave an excellent seminar yesterday about his groups use of computational methods for understanding cancer evolution; and image processing for improved analysis of tissue sections. Florian is one of a relatively rare breed of computational lab heads and is hiring his own wet-lab staff. His seminar was the first place I heard the term "moist-lab".
The rise of the computational research assistant: over the last year or two I have noticed that many wet-lab PI's are hiring a different sort of Research Assistant into their labs: a computational RA. This it is not a surprise given the work those groups are doing, but is a recent change.
Most labs have at least one research assistant, officer, etc., who generally assists in the running of a lab, may often be responsible for maintaining lab stocks and things like cell culture. Research Assistants help labs run smoothly and are often the longer-term members of a group and/or institute. Until recently nearly all Research Assistants worked in the lab and had lab based skills.
But with the amount of data coming out of genomics technologies like microarrays and sequencing increasing so fast a group can quickly become overwhelmed with the volume of data. Simply keeping track of files can be tough.
Hence the rise of the computational RA and the emergence of moist-labs.
Friday, 7 February 2014
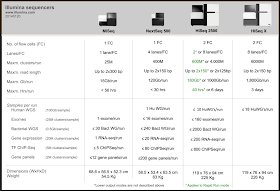
How many samples can you run on an Illumina Sequencer
Illumina now have a sequencer for almost every need: S, M, L & XL. There is a helpful table on their website that give instrument specs and an application-by-application estimate of sample numbers that can be
sequenced in run:
Interestingly gene panel's don't appear on HiSeq 2500. We've used this for targeted sequencing to get phenomenal depth on large numbers of samples to look into heterogeneity in cancer. I'd expect anyone wanting to look deeply into populations would benefit in the same way.
There is also a comparison tool on their website to help you find the right sequencer for your lab: helpful table on their website: interested in RNA-seq for DGE and ChIP-seq in cancer genomics? Here are the results:
Sunday, 2 February 2014
One of our bases is missing: where's the G in NextSeq chemistry
Illumina surprised pretty much everyone with their latest SBS chemistry. I was at an Illumina meeting last week where it was discussed at some length, with many in the audience concerned about possible new error modes due to the drastic change compared to the four-colour SBS used in HiSeq and MiSeq.
I posted an explanation of how the two-colour chemistry works a couple of weeks ago and my initial thoughts remain the same: the chemistry is likely to be an important step forward and Illumina are unlikely to have released it without a lot of confidence in it. So do we need to be particularly concerned about the lack of G signal or the dual-flurophore approach used for A?
Help, my G is missing: argumentum ad ignorantiam, "absence of evidence is not evidence of absence" is the point of view most people seem to have come from. There is concern that a null signal is not the same as a nice bright green spot. But in discussing this with colleagues I was struck by the fact that twenty years ago the lack of signal was the basis for Sanger sequencing working at all. Take a look at the image below, you can read the sequence yourself:
GGTCCGCCACTTCCCGTTAGTC
Anyone who remembers, or actually performed, radioactive sequencing will know what I am talking about. All those lanes to interrogate, reading bases out to a mate while guided by a ruler down the autoradiograph. A genome felt like an impossibility. But the "missing" signal was not an issue, but rather the key to success. 75% of bases in each track had no signal, so what’s all the fuss over the missing G?
But what about comparisons to HiSeq SBS: We'll have to wait for academic groups to sequence larger sample numbers than Illumina have so far produced to get a real handle on how well the SBS chemistry's compare. For now most data is still going to come from four-colour SBS. But in the future who knows. Will two-colour SBS trickle down/across to other platforms? I'm certainly liking the simplification of hardware and looking forward to a more robust instrument. After all most of my groups Tweets are to say how long our queue is or that our HiSeq has broken.
I only ever did radioactive sequencing once during my third year at Uni, it was a two week long exercise, what with development of the autoradiograph, and I got a few hundred base pairs for my project. That was in 1995, nineteen years later and in two weeks an XTen system could generate 640 30x Human genomes.