465 lymphoblastoid cell lines were prepped and sequenced in seven EU labs. TruSeq mRNA seq libraries were sequenced using PE75-100 reads. Each lab ran 50-100 samples but 5 of these were run across all labs. The results were good; in the paper they report very low levels of technical variation, "smaller than the already limited biological variation" and suggest that issues like variation in insert size or GC content between labs can be corrected for. They concluded that large RNA-seq projects could be distributed across labs "given proper standardisation and randomisation procedures", which would have seemed almost impossible in the days of microarrays. "All participating laboratories (must) use the exact same protocols"!
The figure above shows how variable libraries were for (a) total number of reads obtained per sample, (b) mean Q score, (c) mean Q30 length, (d) %duplication, (e) %mapped, (f) %aligned to exons.
A major objective of the study was to evaluate the feasibility of
sharing RNA-sequencing among different laboratories in large consortia
projects and the paper clearly demonstrates that RNA-seq can be highly comparable across labs. As long as samples can be adequately randomised across participating labs then statistical models can be built that will allow smaller technical effects to be removed. This is good for people planning really massive projects. But technology marches on and the HiSeq 4000 can now deliver a project of 500 RNA-seq samples in a single run, making the need to distribute the efforts much less. Whilst the labs in this study have done a great job, they obviously had to work closely to agree on protocols used in the whole process, this communication can be harder than sending everything to one place to get the job done.
They ran a K-mer profiling which appears to be a
useful QC that does not require alignment to a reference genome, I'll look at whether this is something we should be doing here.
Making RNA-seq cost-effective: This project over-sequenced very significantly, and two technical challenges were part of the problem. The first was the use of PE75 vs SE50 sequencing; for RNA-seq differential gene expression several papers have demonstrated that anything longer than SE50 is a waste of time and money. The second issue is one we still wrestle with, sample balance in a pool. In the paper each lab chose how best to pool and sequence samples to get the required minimum of 10M mapped reads or around 5000M reads in total. The variation in read numbers per sample was huge, averaging 58M rather than the 10 required. The authors explained that this "was partly due to differences in the number of samples per lane and partly due to difficulties with equimolar pooling". This over-sequencing resulted in 26,000M reads being generated! As the study used PE75 sequencing (£1200 per lane in my lab); this makes the bill for sequencing about £100,000! We'd typically use SE50 for RNA-seq (£600 per lane in my lab) and if 10M per sample could be obtained this would cost only £10,000.
I'd agree that a big challenge has been to get the balance of samples in a pool as close to 1:1 as possible; we're getting there with our most recent RNA-seq runs, but this is by no means solved. However we always run the entire experiment as one pool (assuming max 96 samples) and run as many lanes as we need, this allows us to keep over-sequencing to a minimum and keep costs down! In figure A from the paper (above) the read distributions are multimodal for all labs suggesting real issues in quantification and pooling. The paper describes issues with different sizes which we almost never see with TruSeq RNA (it's always 270bp).
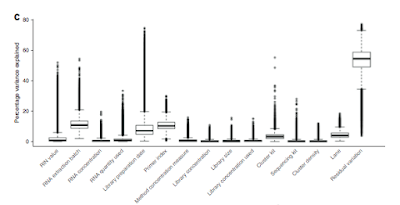
Batch effects revealed: I really liked figure 3d (below) which shows how each of the technical factors considered adds to the variation in the samples. Quite clearly biology is the biggest cause of sample variation, with only RNA extraction batch, library prep date and sample index being factors that may require more careful investigation.