I did not arrive at London Calling until late on Thursday afternoon but what an excellent start I was given: Clive Brown’s update - where ONT are today, and what’s coming tomorrow! The Thursday afternoon, evening and Friday sessions created a palpable buzz in the attendees. It is easy to get carried away with predictions about how soon ONT will become actual competition for Illumina, but it is not clear that this is ONT's main goal. More exciting is the push into uncharted territory, bringing sequencing to individuals and taking it to Mars. I'm excited; but my core facility manager head says, "hold on, you're not getting rid of those HiSeq's just yet"!
Some comments and analysis from the exciting and fast moving world of Genomics. This blog focuses on next-generation sequencing and microarray technologies, although it is likely to go off on tangents from time-to-time
Tuesday, 31 May 2016
Monday, 30 May 2016
Oxford Nanopore's Direct RNA-seq - the killer app for bacterial genomics?
I'll finish my write up of the ONT London Calling event after the bank-holiday weekend but I wanted to get this post out as I think we came up with a killer app for MinION, or even SmidgION on your iPhone: direct RNA-seq from bacterial 16S rRNA. This was an idea I proposed in response to an answer about what one of the speakers was doing with Nanopores (they were working on 16S amplicons). In the ensuing discussion we decided that sequencing the ribosomal RNA directly (see Clive's announcement below) would allow interrogation of phylogenetic relationships and environmental diversity, quickly, and with close to zero bias from sample extraction and library prep.
Monday, 23 May 2016
Increased read duplication on patterned flowcells- understanding the impact of Exclusion Amplification
Next-generation sequencing is fantastic technology and its use has revolutionised our understanding of biology, but it is not perfect, multiple issues occur in every lab from sample extraction through to the actual sequencing. Not all of these are well enough understood to be safely ignored and in this post I'm going to talk about one that I'm trying to better understand right now - duplication of sequences in datasets, and in particular Exclusion Amplicifation duplication on HiSeq 4000.
 |
| Sources of read duplicates in Illumina data - courtesy Illumina 2016 |
Thursday, 19 May 2016
Happy 10th birthday NGS!
NGS is 10...according to the latest Nature Reviews Genetics: Coming of age: ten years of next-generation sequencing technologies. Just by chance I was asked to give a talk to explain how Illumina sequencing works in a technology seminar series being delivered by the Core Heads at the CRUK Cambridge institute, and as part of that I uploaded a slide-deck and created some animations for Twitter to explain how clustering works...I hope you like it.
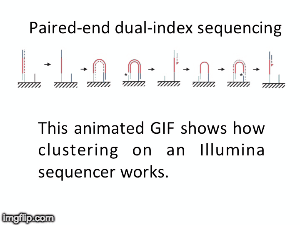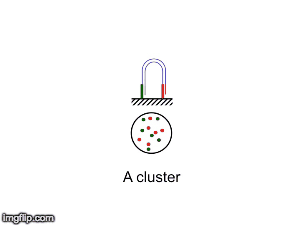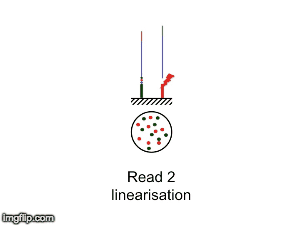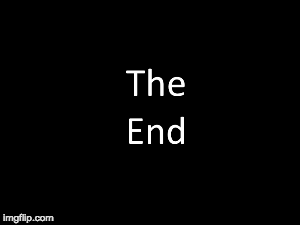 |
| Illumina paired-end dual-index clustering and sequencing |
and here is a "slow-mo" version for people who could not keep up with the frame-rate!
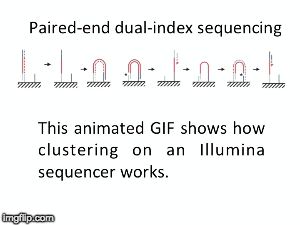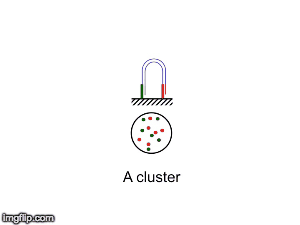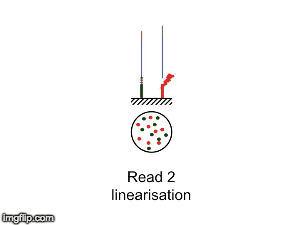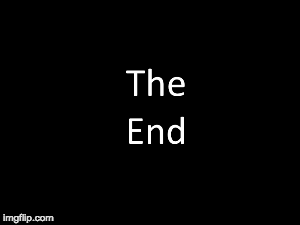 |
Wednesday, 18 May 2016
How will Foundations recent patent announcment affect the cancer testing environment
Foundation Medicine were yestoday granted US Patent 9,340,830
"Optimization of Multigene Analysis of Tumor Samples". This is likely to stir up the can of worms that is tumour testing by NGS and is another patent in a complex landscape. The claims basically cover WGS library prep, exome sequencing, alignment and variant calling. It covers all sorts of mutation calling including SNVs at low freq (5%) and mid-freq (10%) or above, SNPs to assess CNV & LOH, fusions and other structural variants, as well as pharmacogenomic SNPs. It also includes in the test a DNA fingerprint.
Michael Pellini, Foundation's CEO appears to be using some very positive
language in describing the award of this patent, he said "we do
not intend to block the use of methods covered by the patent in patient
testing that may be offered by others". But how much of the patent claim is truly novel and might stand up in court remains to be seen. The basic idea of exome sequencing patients is old hat and Foundation were certainly not the first people to be doing this. The SNP ID of patients is an idea even I'd proposed over four years ago (here & here). But if Foundation's patent makes it harder for others to clamp down on competition that can only be a good thing.
Monday, 16 May 2016
How many genomes can the world sequence per year on X Ten?
Illumina's X Ten was a major announcement. It arguably delivered the "$1000 genome", kickstarted national population genomics as a science, and delivered the final blow to Complete Genomics. It is debatable as to whether Illumina needed to release X Ten at $1000, or with such huge capacity; and it is unclear what the X Ten economics really look like for customers or Illumina themselves, but the world can now sequence an unprecedented number of genomes: about 576,000 per year!
Where are all the X Tens: There look to be about 320 X Ten instruments, I searched online and at AllSeq (great) and Genohub (not so great). Installations include - Baylor College of Medicine (10), Broad Institute (14), CEN4GEN (5), Centre National de GeŽnotypage (5), Core Genetics (?), DKFZ (10), Garvan Institute of Medical Research (10), GENEWIZ (10), Genome Quebec (5), GRAIL (?), HudsonAlpha (10), Human Longevity Inc (20), Macrogen (10), McDonnell Genome Institute (10), New York Genome Center (10), Novogene (10), SciLifeLab (10), Sidra Medical and Research Center (10), SNP&SEQ (?), Theragen Etex Bio (10), Wellcome Trust Sanger Institute (10), WuXi AppTec (10), Genomics England (Illumina) (30), Scottish Genomes Partnership (15), DeCode.
How genomes are actually being sequenced: While each X Ten box can generate one thousand eight hundred $1000 genomes per year it is unclear how heavily they being used. There are very few reports of X Tens being used at capacity and even 80% capacity seems to be optimistic so a large number of those boxes may be sitting idle. The reasons for this are likely to vary from lab to lab but three main factors need to be considered:
- Sample collection: getting enough patients recruited
- The cost of sequencing: finding the cash
- Analysis and interpretation: hiring enough Bioinformaticians
We're just about to send off our first X Ten project for 1000 genomes...tough to do on two HiSeq 4000's!
Friday, 6 May 2016
BaseSpace updated: no more "free" bioinformatics
I've been a big fan of Illumina's BaseSpace since it was launched in late 2011. It was the first truly simple to access and free to use cloud-based analysis infrastructure for NGS data. I've used it a lot - primarily for run monitoring, but also for RNA-seq and Exome QC analysis using the BaseSpace Apps. But the free-for-all analysis Smörgåsbord is ending, and users will be looking carefully at the costs to determine if BaseSpace offers real value when compared to their internal infrastructure.
Tuesday, 3 May 2016
How many reads to sequence a genome?
Last year I posted about the Lander-Waterman equation used to calculate the number of reads needed to sequence a sample. I explained that this general equation (C = LN/G) can be rearranged to allow you to compute the number of reads (N) to sequence a genome of known size (G) with specific coverage (C) and using reads of a specified length (L). Today I finally finished my Calculoid NGS reads calculator to make it easier to access..feel free to use and reuse as you see fit.
Subscribe to:
Comments (Atom)




