Christmas came a little early to my lab with the delivery of two new HiSeq 4000 instruments. It would have been nice if these had come gift wrapped as it is the Christmas season, but alas no. Perhaps next year Illumina can get a little more into the Christmas spirit? I recently contributed to a 2015 review by GenomeWeb and thought I'd give a more in depth view of 2015, and what might be coming in 2016, from my perspective.
Some comments and analysis from the exciting and fast moving world of Genomics. This blog focuses on next-generation sequencing and microarray technologies, although it is likely to go off on tangents from time-to-time
Thursday, 31 December 2015
Wednesday, 16 December 2015
Can't find your primers? Don't bother looking!
PCR primers have become so cheap we had a discussion in the lab today about whether we should simply reorder, rather than try to find some old ones that weren't where we thought they were! This rather glib suggestion made me think about how long I should look before giving up and simply reordering.
At around £0.10 to £0.25 pence per base a pair of PCR primers will set you back £5-10. A post-doc salary is about £16 per hour based on the average salary of about £30,000 per year. So if you can't find your primers in about twenty minutes, give up, place a new order and wait till they arrive.
Wednesday, 9 December 2015
BBC gender equality issues: how do women in science fare?
As part of the BBC's 100 Women Season, they've been putting up some content that allows you to see how equally different countries treat men and women. One of these is the gender equality calculator and according to the Beeb "the UK is among the most gender equal countries in the world". It ranks 18/145 with Iceland top of the league, and Norway, Finland, Sweden and Ireland occupying the rest of the top 5. I was interested to see how equally men and women fared in the sciences, and there is near parity in the numbers graduating from University. However the figures are not so great if you dig into some of the source data...
Thursday, 3 December 2015
Programmable PCR from GNAcode
Fancy being able to run 16 PCR programs simultaneously? The new MiniCube PCR machine from GNAcode does just that. Add qPCR functionality and this machine might improve NGS library prep.
Monday, 30 November 2015
CoreGenomics "Black Friday" sale
The Genomics Core is making space for some new equipment and so we're clearing out some instruments we no longer use. If you are interested drop me a line...sale must end Friday December 4th.
Little Dipper: Automates the washing and processing of Agilent and Illumina microarrays. The Little Dipper controls wash time, agitation, buffer temperatures and drying using an integrated centrifuge. Batches of 1 to 24 slides are processed following manufacturer recommended conditions. Comes with pre-loaded protocols for aCGH, gene expression, miRNA and ChIP-chip applications. Was in ful working order, but not used for over 12 months
BeadArray scanner and autoloader: The BeadArray Reader is a laser scanning confocal microscope system that provides the ability to scan high-density BeadChips. Broken, but the autoloader, including a Twister II arm, were in full working order when last used about two years ago.
HiSeq 2000: not upgradeable to V4 chemistry and unused for two years.
GAIIx paired-end module: Yikes, this was found in a cupboard and has probably been unused for about five years!
Tuesday, 24 November 2015
Communicating with our users: status updates via Twitter
Me and therest of the Genomics Core team (past and present) put quite a bit of effort into communicating with our users. This ranges from simple communications about when a
project will be complete, or a sample might be run; to more complex
communications like experimental design discussions. The group website had a major update two years ago and we started a group blog, we have an email HelpDesk, and have just added a new Twitter account specifically to inform users how long our Illumina sequencing queue is: @CRUKgenomecore puts out live Tweets directly from our Genologics Clarity LIMS.
Friday, 20 November 2015
Ten X updated...rare disease anlaysis made easier?
We're about to start a pilot work project with 10X Genomics so I put together an update to share internally and thought it would make a good post here too...I've been thinking about how 10X might be used in cancer and rare disease, as well as non-human genomes. The NIST results described below are particularly interesting as the same sample has been run on all of the major long-read technologies: 10X Genomics, Moleculo, CGI LFR, PacBio, BioNano Genomics and Oxford Nanopore; as well as both Illumina and SOLiD mate-pair. Enjoy.
Monday, 16 November 2015
Comparing reference standards for NGS cancer genomics
Cancer genomics is rapidly moving away from pure discovery to a much more translational perspective. An important area to consider, as the tools being used and developed in research labs are deployed to a more clinical setting, is an understanding of the reproducibility, sensitivity and specificity of these tools. A big help in this is the availability of high quality reference materials, which are likely to form an important part of any clinical lab QA program. Several companies sell reference standards but the most interesting in the context of this post are those from Horizon Discovery, AcroMetrix and Seracare; each of which is briefly reviewed below.
 |
| Horizon Diagnostics approach is one of several |
Wednesday, 4 November 2015
How many liquid biopsies per year by 2020: in the UK it might be well over 1 million
I've been doing some work looking at CRUKs cancer incidence statistics and this is what sparked the idea for this post. There are just under 350,000 new cancer cases in the UK each year.
Although ctDNA can be found in many/most tumour contexts (Bettegowda et al 2014), it may not be used universally. I'll assume that in all common
cancers ctDNA sequencing, a "liquid biopsy"(not sure how Cynvenio got there first with the trademark - smart move), will be used as a first-line and/or
follow up test run at least once per year per patient on average - by 2020.
Assuming that patients will live for 10-20 years with their disease this
works out at 1.75 million to 3.5 million tests per year. I'm sure people
are doing much better modelling than me but by any measure this is a lot
of sequencing tests to run!
Sequencing can seem
difficult, however to be able to sequence
all those tests someone's got to make 1,750,000+ libraries! An
automation
platform capable of processing 96 samples in 24 hours would need to run
for fifty years! Or fifty labs would be required just to keep up
with demand. Even at just £25 per sample for an amplicon based test
(no-one is offering a test at this price today) the costs would be
almost
£50M-100M. This assumes that the cost of collecting samples is zero, and
I'll totally ignore Bioinformatics which I think will disappear for
simple analysis by 2020*.
Right now there are no library
prep methods that truly allow you to go
from sample to sequence in one day. Innovation here is going to be
vital, and I suspect will become more and more the focus for investment.
The company that can come up with a fast and robust method, and sell it
to someone like the NHS in large volume is going to come out on top.
Might we even get to a similar situation as with forensics where only a
couple of tests are internationally recognised, making the sharing of
data much easier?
NIPT is being rapidly adopted partly
because the tests have been rapidly defined - the coverage required for a specified sensitivity/specificity is known. Somatic sequencing is tougher
due to lack of clarity of the sensitivity required to have a clinical impact. Clinical trials are happening now but it might remain an open question for a while as to whether you should swap to a different drug, e.g. EGFR inhibitor, or combination, when e.g. T790M gets to 1%, 10% or 50% mutant allele frequency.
Worldwide there are over 14 million new cancers per year, if the logic above translates then the number of tests climbs fast - maybe 150M liquid biopsies per year.
*Whilst the bioinformatics challenges are huge right now I do believe that we'll have well developed pipelines that can call mutations and generate clinical reports with minimal effort for "simple" cancer panels by 2020.
Monday, 2 November 2015
1000 citations and counting
I updated my Google Scholar account today and saw that a paper I was a co-author on has had over 1000 citations: The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups! This now puts it up near the top of Natures paper mountain, although still behind Watson and Crick's 1953 paper. The main reason I am writing about this here is not self-promotion (there were 33 co-authors, and I was part of the METABRIC group - itself a collection of around 80 other co-authors), but rather a chance to discuss how we might approach this with RNA-seq as opposed to the Illumina microarrays we used.
METABRIC: The Nature 2013 METABRIC paper (Molecular Taxonomy of Breast Cancer International Consortium) described the analysis of over 2000 breast cancers using copy-number profiling (on Affymetrix SNP6.0 arrays), and gene expression (on Illumina HT12 arrays). It highlighted the molecular complexity of breast cancer in a way not previously revealed and stratified cases into one of ten distinct molecular clusters (IntClusts), that had different outcomes. Read the paper, or the CRUK blog for more info.
Microarrays not RNA-seq: The METABRIC paper was published in 2013, but the ground work began in 2006, in fact this project was a deciding factor in my taking on the role of Genomics core facility head at CRUKs Cambridge Institute. Back in 2006 only a very small number of people were working with NGS and no-one for RNA-seq (the first RNA-seq paper was not published until 2008 Nagalakshmi et al: Science). Microarray gene expression analysis was the state-of-the-art and we had invested in Illumina's beadarray technology rather than the more common Affymetrix chips).
We planned the microarray processing incredibly carefully, it took months to decide on how samples would be randomised across plates, how extractions would be performed, etc. In the end we processed around 2000 arrays in the first round. To increase the quality and comparability of data across the project the whole lot were processed in one six-week batch (we did no other array work during this time), and all the samples were prepped as labelled cDNA by one person*. An additional 900 arrays were completed as follow up, some repeats, some new samples.
Illumina's HT12 array really made the microarray processing much simpler. 12 samples were hybridised on one array with a full 96-well plate being run each day. All arrays were run as a batch and scanned overnight. I'm sure we could have got away with an HT24 format, and if this had been available then the costs might still be competitive with RNA-seq.
Doing it all again with RNA-seq: We are just completing a 600 sample RNA-seq project using our Agilent Bravo and Illumina TruSeq stranded mRNA-seq kits. The processing is pretty straightforward and we can go from RNA to a normalised pool of 96 samples (actually 88 but don't ask why) for sequencing in about a week. Our normal project size has increased each year, and although 600 is still large, we usually process a batch of RNA-seq samples every week or two.
If we were to run METABRIC again I'd like to increase the amount of automation and the number of indexes to 384 or even 1536. With a kit cost of about £30 ($50) per sample, and 20M single-end 50bp reads per sample costing about £50 ($80), the whole project could be completed pretty quickly and for just £160,000 ($250k).
But what really amazes me is that we can sequence 2000 samples for high-quality and sensitive mRNA gene expression in a little over three weeks on our HiSeq 2500, probably even quicker on the newer HiSeq 4000. We'd also be able to go back to specific samples, or sub-groups, and sequence deeper to look into alternative splicing.
RNA-seq continues to develop with newer kits, better analysis of non-coding transcripts and development of methods to call mutations and isoforms. Recent advances like the SEQUEL from PacBio offer the chance to directly count isoforms rather than infer them from short-reads. And who knows how soon RNA-seq on the MinION will be a reality for the masses?
Microarrays not CNV-seq: We processed all the samples for METARBIC copy-number analysis through Aros in Denmark using Affymetrix SNP6.0 arrays. But we have been using low-coverage whole genome sequencing for a number of years now as part of our exome capture pipeline. Pre-cap libraries are pooled for very low coverage sequencing (about 20M reads) and analysed with QDNA-seq (Scheinin et al: Genome Research). We spent probably over £1M on Affy SNP6.0 arrays versus about £100,000 for sequencing . The sequencing workflow is similar to RNA-seq and we'd be able to zoom into specific samples, or sub-groups, to get a better handle on those that are driven by copy-number changes. We'd also be able to detect other structural variants.
Big projects can present many challenges and I am sure the authors that prepared the METABRIC paper had to defend against the use of arrays just as RNA-seq was starting to gain traction. It can be difficult to decide which technology to use, I am certain that we chose the right things for METABRIC, but that choice was one made after lots of discussion. If you're planning a big project, or even a very small one, take the time to think about the methods your using, their biases, and yours, before starting the lab work.
*Many thanks should go to Michelle who did all the sample prep. She was pregnant at the time and left shortly afterwards to have her baby, and for a well earned rest! Michelle left the lab in 2014 and now works at Iniviata.
Tuesday, 20 October 2015
Why do you read science blogs: a research project funded by Experiment
This is not wholly a direct request for feedback on the Core Genomics blog! Dr.
Paige Jarreau from LSU is surveying science blog
readers about their social media habits and perceptions of the science
blogs they read. Her project is funded partly through Experiment: the kick-starter of the experimental world. Her project brought this platform to my attention and it looks pretty cool (see the latter part of this post).
Tuesday, 6 October 2015
X Ten: now available for non-Human
Finally. Today Illumina announced that X Ten users can perform whole-genome
sequencing of non-human species. Does this mean exomes and RNA-seq are on their way to X Ten? Or that an X One is going to be announced at JP Morgan? I might just hold off on buying that shiny new HiSeq 4000 until this is a bit clearer.
For small to medium labs the X Ten and even X Five were the stuff of dreams and nightmares. Dreams of what would be possible with this technology; nightmares of what might happen to smaller labs.
The dreams turned out to be true and the nightmares have pretty much gone away. But the rapid developments from Illumina continue to be difficult to keep up with. I only wrote about the performance of HiSeq 2500 V4 chemistry a little over a year ago, and it looks like we'll ditch it in favour of HiSeq 4000 in 2016.
The link on Illumina's webpage does not appear to be live just yet but I'm sure we'll hear more in the next couple of days.
PS: If your Wheat, Horse, Whale, etc, etc, etc genome sequencing grant just got approved you can look forward to a nice slush fund. Time to buy that Apple watch maybe?
PS: If your Wheat, Horse, Whale, etc, etc, etc genome sequencing grant just got approved you can look forward to a nice slush fund. Time to buy that Apple watch maybe?
Friday, 2 October 2015
Pub-Bed: beds, not papers
Would you stay at the home of another academic you had some loose connection with? Could the Airbnb model be successfully applied to help find accommodation for scientists travelling to meetings, visiting another lab, or even for longer sabbatical stays? I'm not sure but Pub-Bed was born from an idea I cooked up on the train.
Thursday, 1 October 2015
The new Pacific Biosciences sequencer
PacBio announced a baby RSII yesterday, which should be in the shops just in time for Christmas! The Sequel System (sounds like SequalPrep from Thermo for PCR cleanup) sounds like a big advance on the enormous RSII. Most aspects of the sequencing work flow are unchanged. Sequel has been developed as part of the collaboration with Roche to develop a diagnostics instrument, milestone payments of $20-40M are expected on the back of this.
- $350,000 for Sequel (versus $1,000,000 for RSII)
- Seven times more reads than RSII
- 1/3rd the size and weight (so only 2000 or so MinIONs will fit inside)
- New SMRT cells with 1,000,000 ZMWs compared to RSII's 150,000
Tuesday, 29 September 2015
Do you have a HiSeq 2500 V4 to sell
My lab is looking to put an additional HiSeq 2500 v4 in place, Illumina do not have any (refurbs) to sell in the US or Europe so I thought I'd post here to see if any Core Genomics readers have an instrument they are looking to retire? If we can offer you more than Illumina's trade-in on a HiSeq 4000 then we might be able to come to some arrangement!
Drop me an email at James.Hadfield@cruk.cam.ac.uk if you might be able to help.
Friday, 25 September 2015
Plagiarism alert via Twitter
I built several TwitterBots last year to scrape papers on PubMed (thanks again to Casey), these have turned out to be really useful in alerting me to new work but last night I got a very interesting Tweet from @MattiasAine...
Wednesday, 23 September 2015
Rubicon Genomics webinar on making cfDNA/ctDNA NGS libraries
Rubicon Genomics have a webinar on Creating Cell-free DNA Libraries with ThruPLEX Plasma-seq that was broadcast on September 17th 2015. The new PlasmaSeq kit has a very simple, 3-step, 15 minutes hands on time, single-tube workflow.
Dr. Kamran Shazand, Rubicon Genomics' Director of Applications presented
an intro to cell-free DNA - generally 170bp fragmented DNA, which is
the result of degradation by apoptocic enzymes, and that can appear in
dimeric and trimeric nucleosomal fragments. Rubicon's earlier chemistry ThruPLEX was use in 2012/12 to sequence foetal and tumour genomes and it has a recommended input of 1-30ng of DNA.
The new PlasmaSeq kit seems to make very nice libraries. Internal data from Rubicon showed better performance against NEBnext and KAPA Hyper, with high diversity, low PCR duplication rates and good GC coverage. Most interestingly Rubicon have built a target capture pipeline using their PlasmaSeq kits, Agilent SureSelect and IDT blockers allowing exome sequencing from as little as 1ng of input DNA.
Methods: optimised DNA repair and ligation of stem-loop adapters, PCR to extend, cleave and amplify the library. You should end up with a major peak at 310bp for cfDNA libraries.
Rubicon also talked about their work looking into how molecular indexes night be added to allow removal of PCR duplicates to PlasmaSeq - -send them your comments if you are interested.
Disclosure: I am a user of Rubicon library prep technology; but also Illumina, NEB, Enzymatics, and many others too! I am an author on the ctDNA paper from 2012 mentioned above and in their webinar. I am on the SAB of Rubicon.
Research Professional: making finding funding easier
I'm incredibly lucky because I don't usually need to write grants. My lab is core funded by CRUK to provide genomics services to the CRUK- Cambridge Institute. However I do like to keep an eye out for opportunities to bring funding in for specific projects.
Our very good grants team pointed me to a tool that the University subscribes to *Research Professional. Once logged in I can easily search for new funding opportunities, if you've never used it it could become one of your favourite websites.
Research Professional began as a (print) “newspaper for the research world" in 1994, and moved to the web in 2001. It make accessible the opportunities and information on research grants to registered users and may institutions have purchased access. If your host Institute does subscribe then you should certainly take a look! You can search very easily and follow links directly to funders websites. The site generally provides a good summary of funding criteria so you can quickly weed out opportunities that you will not be eligible for.
Our very good grants team pointed me to a tool that the University subscribes to *Research Professional. Once logged in I can easily search for new funding opportunities, if you've never used it it could become one of your favourite websites.
Research Professional began as a (print) “newspaper for the research world" in 1994, and moved to the web in 2001. It make accessible the opportunities and information on research grants to registered users and may institutions have purchased access. If your host Institute does subscribe then you should certainly take a look! You can search very easily and follow links directly to funders websites. The site generally provides a good summary of funding criteria so you can quickly weed out opportunities that you will not be eligible for.
Monday, 21 September 2015
Better ChIP, requires better antibodies
ChIP-seq can only really work if you have a good antibody for the protein of interest. Any non-specific binding is going to add noise to your data making motif finding, differential binding analysis difficult or even impossible. Back in 2008 Mathias Uhlén's group at the Royal Institute of Technology, Sweden, published a study that showed how less than 50% of antibodies tested were specific to the target.
Wednesday, 9 September 2015
#GenSci15 plenary session and wrap up
Sorry about the accidental posting...here's the final version.
Innes Barroso, Head of Human Genetics at Sanger Institute "Genomics and Metabolic Disease – what have we learned so far"
Type 2 diabetes, many patients controlled by table some on insulin, it is a very complex disorder, both genetic and environmental effects (particularly obesity, diet, exercise). Incomplete understanding of Type2 diabetes genetics, extended families are hard to obtain due to generally late onset. Progress has been slow, early studies failed to find robust loci that replicated across studies. However as power of GWAS studies improved six loci were found outside previously suspected candidate genes, but with small effect. Analysis of extremes can be very powerful i.e. young, lean patients vs old obese controls (similar approach being pursued in smoking with some success). Development of genotype imputation and meta-analysis had big impact on how GWAS could impact Type2 diabetes genetics. But still lots of "hidden heritability", drove development of custom GT chips e.g. Metabochip and UK Biobank Axiom array.
#GenSci15 day3 clinical genomics session
Andrew Beggs is chairing the clinical genomics session – this is why I am here, cool stuff!
Interesting that the Tolstoy quote from Pippa Thomson's talk (see below) "Happy families are all alike; every unhappy family is unhappy in its own way" came up, as it could be argued that the Anna Karenina principle probably applies to the clinical use of genomics right now: a deficiency in any one of a number of factors dooms it to failure. Clinical genomics can only have a successful impact if every possible deficiency in the study has been avoided i.e. clinical and family history, quality of exome coverage, issues with alignment etc, etc, etc,
Tuesday, 8 September 2015
#GenSci15 day 2
Day 2
Single cell was popular, but were all crammed over in the Arts building somewhere close to Solihull. Nick forgot to organise a coach so we had to walk, I guess we're building up an appetite for the street food tonight! Bill Hanage asked if Nick could sequence some of the street food just in case - perhaps next years conference pack can come with a stool collection kit for some crowd-sourced science: "the differential impact of conference attendance on gut microbiota of PhD students, post-docs and PIs"?
Single cell was popular, but were all crammed over in the Arts building somewhere close to Solihull. Nick forgot to organise a coach so we had to walk, I guess we're building up an appetite for the street food tonight! Bill Hanage asked if Nick could sequence some of the street food just in case - perhaps next years conference pack can come with a stool collection kit for some crowd-sourced science: "the differential impact of conference attendance on gut microbiota of PhD students, post-docs and PIs"?
#GenSci15 day 1
I'm at the 6th UK Genome Science conference in beautiful and vibrant Birmingham. There is a good attendance and the place is full with a real buzz about new technologies (more about that in a sec), the work people are doing and the freebies on the stands!
I'm not going to attempt to round up each and every day but I'll probably post tomorrow and Wednesday on my favourite bits. Here's what happened yesterday...
Daniel MacArthur: Mass Gen, Broad, Harvard spoke about the lessons from analysis of 60,000 human exomes. He was making the point that "making sense of one genome requires thes of thousands of genomes." In the ExAC (exome aggregation consortium) they've gathered almost 100,000 exomes which have then been consistently analysed as a single batch to produce a single VCF. Dan showed that by doing this they were able to remove noise from rare disease analysis - and although I'm not sure about the impact on cancer exomes it is likely to make it easier to filter variants from those datasets too.
Tuesday, 1 September 2015
S is for Sequencer: the new instrument from Ion Torrent
Today Ion Torrent launched their newest sequencers the S5 ($65k or $150k with the S5 XL, which includes more compute). Ion Torrent was exciting technology when we first heard about it and I'm disappointed it never quite lived up to the promise, and that it never competed against Illumina in the space I cared about. The new systems do not change my disappointment, but they may increase the competition for Illumina in the amplicon sequencing space. In my lab we're still running lots of amplicons using Fluidigm and HiSeq and I don't think we'll be buying an S5 anytime soon. However labs looking to set up clinical (or other) tests on standard panels might see the S5 as a realistic alternative to a MiSeq. But Ion need to deliver on this visibly and believably, I know lots of labs that have stopped using their PGMs, although I know a few that have stopped using their MiSeq's too!
The YouTube video (above) offers a very marketing-heavy overview (where is the S5 vs MiSeq video). Ion have focused very clearly on amplicons and delivering results as quickly as possible at reasonable cost. The S5 XL includes additional compute for processing to speed up this data delivery in an automated manner.
Like PGM and Proton before, S5 offers multiple chip configurations that allow you to run fewer samples without sacrificing per sample costs (too badly). The 520/530 chips are equivalent to the PGM offering 200-400bp runs with 5 or 20M reads per run. The 540 chip only
offers 200bp runs but with 80M reads. All with 2 to 4 hour run times and analysis completed in as little as 5 hours.
S5 and Ampliseq: An open letter from Mark Gardner (Ion's GM) at the Behind the Bench
blog explains the desire to continue the democratisation of sequencing.
The aim was to make targeted NGS easy for anyone to work with, and to
deliver the best value benchtop sequencer. As the S5 system is very obviously targeted to amplicons I thought I'd highlight what Ion say in their literature about the numbers of samples per run - using the Ion AmpliSeq Cancer Hotspot Panel v2 (50 genes), 16, 48 or 96 samples can be run on 520, 530 or 540 chips respectively.
You'll need the Ion Chef in your lab to go from DNA to data with less than 45 minutes of hands-on time; otherwise you'll have to manually prepare libraries and templates for chip loading.
Data analysis can be completed automatically within 5 hours.
This does offer a tempting solution for people sequencing panels, but it may be limited in what else is realistically possible on other (Illumina) sequencers. The low-cost is going to be attractive, but only to a point. If Ion can deliver a truly end-to-end solution then they may be onto a winner.
AllSeq got their first with their digging around on Google, and Keith at Omics Omics got a sneak peek, "Ion have identified a profitable market segment...fast, targeted sequencing -- and is going full bore in this area". Keith points out that the new chips are not compatible with PGM or Proton, "the concept that all upgrades are encased in the consumable is long dead".
He goes into some depth about the competition for these new
instruments; MiSeq (takes four hours to deliver 1x36bp); GnuBio,
Genia/Roche and QIAGEN have gone all but silent (expect more from Qiagen
very soon), and ONT's MinION (possibly).
See it yourself on the Ion Torrent world tour. The UK Genome Sciences meeeting does not feature on the list of upcoming conferences where Ion will present data - I am sure there's still room. You can catch them in London on the 10th.
I'll
be interested to start talking with users at AGBT in February. Whether
the S series will take off where PGM and Proton did not may partly
depend on whether PGM and Proton users get a good trade-in.
Sign up for Ion World Tour location near you >
Sign up for Ion World Tour location near you >
Sign up for Ion World Tour location near you >
PS: If I wanted to trade-in a HiSeq 2000 for amplicon work what would Ion offer me?
Tuesday, 25 August 2015
Normalisation made simple...please?
We do lots of DNA, RNA and NGS library normalisations mostly by hand after calculating dilutions in Excel. It works well but is cumbersome and prone to error. We're fortunate to have an Agilent Bravo robot and are now automating more of this with excellent results. The most obvious impact has been on NGS library pooling, below is our most recent run where the variability on all but 9 libraries is pretty much perfect.
However automation is a pain and it would be great if there were a solution for normal labs. We've discussed an idea with pipette companies in the past but no-one appears to offer anything yet so I thought I'd suggest it here as well. If you've seen something on the market please do comment and let me know (Sartorius are almost there with the eLine pipette see link below).
If you like the idea follow up with you favourite pipette rep and lets see if someone can make one!
The background to the idea: normalisation can be done by combining a standard volume of sample with a know volume of water or diluent. Any lab trying to do this manually is almost certainly taking concentration measurements from PicoGreen or qPCR and working out how much water to add to each well of a 96well plate with sample already added.
The idea: We
use repeat pipettes for may applications and there do exist
programmable versions. Repeat pipettes are great for adding the same
volume of a master mix to all wells of a plate and several companies
make them. We use the slightly older Eppendorf Stream but Gilson, Sartorius and others exist.
A programmable repeat pipette that can accept a TXT file. The upload of a TXT file after calculating the amount of water to add to each well, and after specifying pipetting across rows or down columns, would allow the user to complete normalisation in as few as 9 pipetting steps: 8x12 channel 5ul transfers from the source plate to a normalisation plate, and 1x automatic repeat pipette dispense. "Simples" as the Meerkat says.
Friday, 21 August 2015
Reproducibility of RNA-seq
The GEUVADIS Consortium published a study looking at reproducibility in RNA-seq in Nature Biotech in 2013 and I only just saw the paper. I thought that readers of this blog would be interested so take a look: Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories.
465 lymphoblastoid cell lines were prepped and sequenced in seven EU labs. TruSeq mRNA seq libraries were sequenced using PE75-100 reads. Each lab ran 50-100 samples but 5 of these were run across all labs. The results were good; in the paper they report very low levels of technical variation, "smaller than the already limited biological variation" and suggest that issues like variation in insert size or GC content between labs can be corrected for. They concluded that large RNA-seq projects could be distributed across labs "given proper standardisation and randomisation procedures", which would have seemed almost impossible in the days of microarrays. "All participating laboratories (must) use the exact same protocols"!
The figure above shows how variable libraries were for (a) total number of reads obtained per sample, (b) mean Q score, (c) mean Q30 length, (d) %duplication, (e) %mapped, (f) %aligned to exons.
A major objective of the study was to evaluate the feasibility of
sharing RNA-sequencing among different laboratories in large consortia
projects and the paper clearly demonstrates that RNA-seq can be highly comparable across labs. As long as samples can be adequately randomised across participating labs then statistical models can be built that will allow smaller technical effects to be removed. This is good for people planning really massive projects. But technology marches on and the HiSeq 4000 can now deliver a project of 500 RNA-seq samples in a single run, making the need to distribute the efforts much less. Whilst the labs in this study have done a great job, they obviously had to work closely to agree on protocols used in the whole process, this communication can be harder than sending everything to one place to get the job done.
They ran a K-mer profiling which appears to be a
useful QC that does not require alignment to a reference genome, I'll look at whether this is something we should be doing here.
Making RNA-seq cost-effective: This project over-sequenced very significantly, and two technical challenges were part of the problem. The first was the use of PE75 vs SE50 sequencing; for RNA-seq differential gene expression several papers have demonstrated that anything longer than SE50 is a waste of time and money. The second issue is one we still wrestle with, sample balance in a pool. In the paper each lab chose how best to pool and sequence samples to get the required minimum of 10M mapped reads or around 5000M reads in total. The variation in read numbers per sample was huge, averaging 58M rather than the 10 required. The authors explained that this "was partly due to differences in the number of samples per lane and partly due to difficulties with equimolar pooling". This over-sequencing resulted in 26,000M reads being generated! As the study used PE75 sequencing (£1200 per lane in my lab); this makes the bill for sequencing about £100,000! We'd typically use SE50 for RNA-seq (£600 per lane in my lab) and if 10M per sample could be obtained this would cost only £10,000.
I'd agree that a big challenge has been to get the balance of samples in a pool as close to 1:1 as possible; we're getting there with our most recent RNA-seq runs, but this is by no means solved. However we always run the entire experiment as one pool (assuming max 96 samples) and run as many lanes as we need, this allows us to keep over-sequencing to a minimum and keep costs down! In figure A from the paper (above) the read distributions are multimodal for all labs suggesting real issues in quantification and pooling. The paper describes issues with different sizes which we almost never see with TruSeq RNA (it's always 270bp).
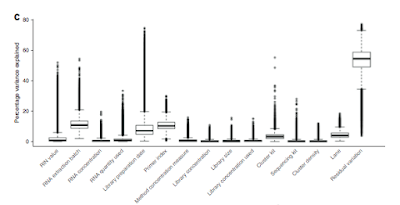
Batch effects revealed: I really liked figure 3d (below) which shows how each of the technical factors considered adds to the variation in the samples. Quite clearly biology is the biggest cause of sample variation, with only RNA extraction batch, library prep date and sample index being factors that may require more careful investigation.
Saturday, 25 July 2015
Thursday, 23 July 2015
Cell-free DNA trisomy 21 tests kick ass
NIPT
for Down's Syndrome and other chromosomal abnormalities is taking off. A
colleague of mine recently had an Ariosa test, paid for privately, and
reported real satisfaction with the process. Lin Chitty at UCL Institute of Child Health and Great Ormond Street Hospital recently reported on a model-based analysis of NHS costs and outcomes of NIPT for Down's syndrome.
This suggested that NIPT was cost effective if offered at around £50
per test (compare this to the £500-£1000 privately). NIPT is not my area
of expertise but I've been watching it as technological developments
have often been a little in advance of cell-free DNA work in cancer.
Possibly millions of tests have now been performed. NIPT is being rolled out to patients across the globe at an amazing rate compared to the introduction of other diagnostic tests, and the NHS is getting in on the game. The number of companies offering tests is growing and so are the litigation's. Most recently Illumina filed a new patent infringement suit against Ariosa claiming their Harmony NIPT test infringes a patent for “Multiplex Nucleic Acid Reactions” (one of the patent holders is ex-Illumina, and was an author on the Ariosa paper discussed below). NIPT commonly tests for trisomy 21 (Down’s Syndrome), trisomy 18 (Edwards’ Syndrome) and trisomy 13 (Patau’s Syndrome) and most tests are NGS based, Ariosa's test is array based. You can get an NGS-based NIPT test from ThisIsMy for just £320, tests in North America are as low as £200. Tests are available from: Ariosa Harmony, BGI NIFTY, Genesis Genetics Serenity (Verifi), Illumina Verifi, Natera Panorama, Premaitha IONA, Sequenom MATERNIT21.
What do you want to be when you grow up?
The MRC have a nice career mapping tool: Interactive career framework which allows biomedical researchers to navigate through different options to see how they might get where they want to.
I'd like to think of myself as a technology Specialist Director: "an individual with technical expertise / specialist skills useful beyond their own specific group" - what are you?
I'd like to think of myself as a technology Specialist Director: "an individual with technical expertise / specialist skills useful beyond their own specific group" - what are you?
Wednesday, 15 July 2015
How should I store my NGS data: disc, tape or tube
Genomics has recently been singled out as one of the largest data headaches we face. As we move to sequencing people multiple times, start newborn genome sequencing programs and increase our use of consumer genomics the amount of data goes up and up. Our GA1 generated 1Gb of data in about 11 days. Today our HiSeq 2500 puts out 1TB in 6.
We're currently storing our data on disc for up to six months. After this we either delete it or archive it onto tape (although Ive no idea if we ever try to get it back off the tapes). A while back people used to talk about the storage being more expensive that a rerun, and I wonder if we are getting even closer to that point, especially if you try to grab the data off a tape in a secure storage facility.
I've always liked the idea of storing libraries and we have all 10,000 that we've run safely stored at -80C. These tubes take minimum space and most could be rerun today at a fraction of the cost from a few years ago. I am now wondering if we should go for an even greener solution and start the long term storage on Whatman cards (available from CamLab and others). A small number of cards could store almost everything we've ever run!
Is anyone doing this?
Tuesday, 14 July 2015
An example of how fast NGS develops
Illumina have discontinued the version 1 of the NextSeq chemistry. Launched in January
of last year the NextSeq was a revolutionary new sequencer, although not everyone was an immediate fan. The V2
chemistry was launched just before AGBT and the data certainly looked a
lot closer to the quality we expected from the longer-lived 4-colour SBS
chemistry. The V1 discontinuation notice arrived in my InBox today,
just 18 months after the NextSeq launch.
That's not much longer than the shelf-life of a kit!
Monday, 13 July 2015
Your genome for under £2000
Illumina have a new offer on their Understand Your Genome (UYG) program that means you can get your genome sequenced, analysed and clinically interpreted for under £2000.
Interested? Then there are a few requirements, mainly that you give informed consent and get a doctors prescription for the test. Your DNA is sent to Illumina's own Clinical Services Laboratory, CLIA-certified since 2009. The results will be reported to you at the first day of the ASHG meeting in Baltimore. Samples need to be with Illumina by July 31st giving them 67 days for sequencing and analysis.
You'll get back results on 12 genes important in pharmacogenomics, and hundreds of genes implicated in human disease. However you'll need to discuss any "medically significant results" with your GP, and you can ask not to receive some data back.
You'll get back results on 12 genes important in pharmacogenomics, and hundreds of genes implicated in human disease. However you'll need to discuss any "medically significant results" with your GP, and you can ask not to receive some data back.
Sounds like a pretty good bargain given you'd need to sequence 50+ genomes to get close the $1000 genome from an X Ten provider. I'm not sure if you'll find out how much Neanderthal you're carrying around?
PS: If anyone fancies crowd-sourcing a Hadfieldome drop me a line, or my PayPal account is...
Thursday, 9 July 2015
Exciting developments in Pancreatic Cancer
A paper just published in Nature Communications describes a molecular analysis of Pancreatic Cancer by tumour exome and ctDNA targeted sequencing. The results showed enrichment of mutations in known PaCa associated genes, and identified clinically actionable mutations in over 1/3rd of patients.
Tuesday, 7 July 2015
MinION for 16S rRNA-seq
Researchers in the group of Yolanda Sanz in Spain deposited a preliminary MinION study describing the bioRxiv of 16S rDNA amplicon sequencing from a mock microbial community composed of genomic DNA from 20 different bacterial species (BEI Resources).
Experimental workflow: 1.5kb amplicons were generated from 16S rRNA gene sequences for 20 different species present in the mock community using a universal PCR. Amplicon library prep was performed using NEBNext End Repair Module and NEBNextdA-tailing module to prepare blunt-end amplicons for adapter ligation. Sequencing was on an R7.3 flowcell with a 48 hour run with an additional library loading at 12 hours. Read QC and conversion to FASTA was performed with poretools and poRe. And they also discussed the "What's In My Pot" Metrichor workflow demonstrated at ASHG, showing that MinION can deliver real-time analysis making the "run until" mode an attractive one for some applications.
Results: Most reads were around the size of the amplicons (median 1.1kb) but they did see some very long reads (max 50kb) speculated that these were amplicon:amplicon ligation products but they were not able to align these. In the reported analysis they filtered the MinION reads to keep only those that were 100bp-2kb, retaining 97% of data (3,297 reads). The 2D reads were discarded due to having "a detrimental effect of 2d reads in the quality of assembled sequences"; given that others are reporting that 2D reads are what we should be aiming for I'd have liked some explanation as to why these were so bad.
They were able to reconstruct more than 90% of 16S rRNA gene sequences for all organisms included in the mock community. However quantitation data was less convincing and the data in figure 1 do make me wonder how applicable this method/tech might be to quantitative or longitudinal analysis. The mock community contained equimolar ratios of 20 rRNAs yet results varied by up to 100 fold, although the authors only considered a coverage bias was present if it was more or less than 10 fold from the expected value. I have no idea what allele frequency is usually tolerated in these kind of experiments.
MinION currently generates a relatively low per-base sequencing accuracy but the additional read length here helps in resolving 16S rRNA to a species level. A nice dynamic figure would be an evolutionary tree of species showing how resolution changes with accuracy and length would have been great!
MinION methods are developing rapidly and I wonder how long will it be before full length 16S rRNA is reported?
Subscribe to:
Posts (Atom)